
About Us
Our group is working on understanding the role of water in protein folding, hydration, dynamics, and the landscape of intrinsically disordered proteins (IDPs). We are using a combination of molecular dynamics simulations and machine learning to study these complex and dynamic processes. The goal of our research is to gain a better understanding of the molecular mechanisms of protein folding and to develop new methods for predicting the structure and dynamics of proteins.
The Essence of Life: Hydration Water in Proteins:
Ongoing Work:
Currently working in role of hydration water of different biological systems as well as water mediated long range interaction
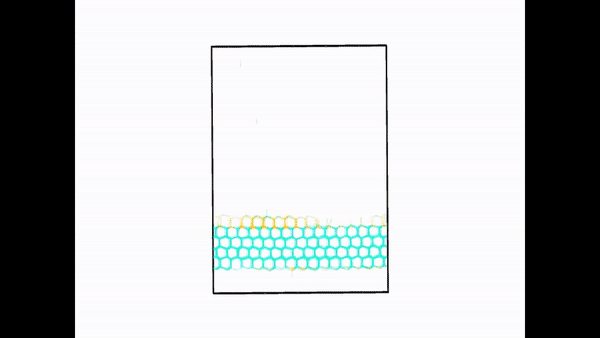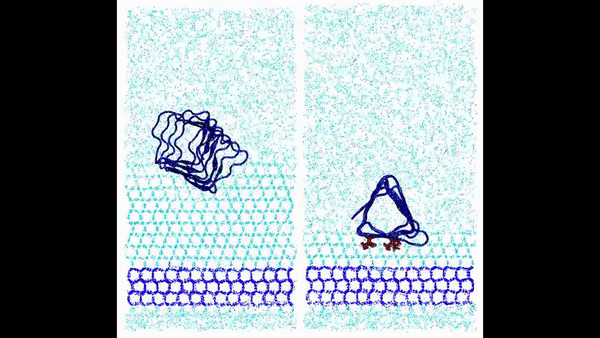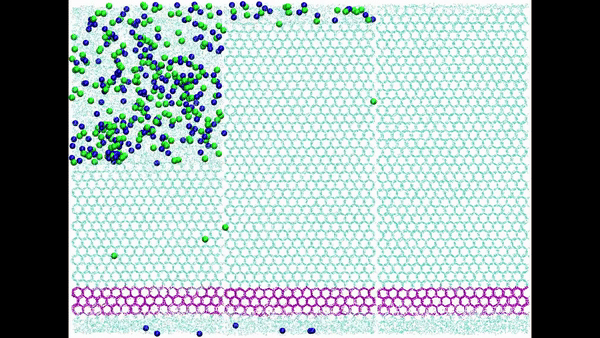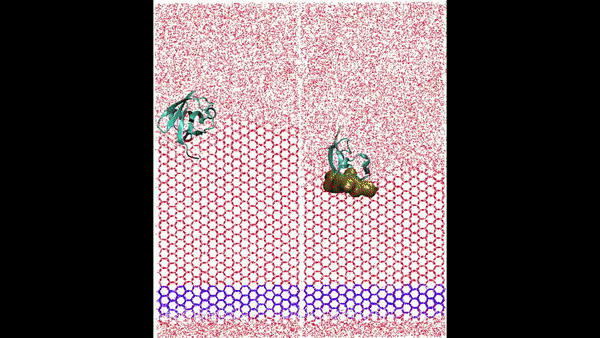
Hydration water is essential for the stability and function of biological molecules like proteins and enzymes. It deeply influences their structure, flexibility, and efficiency. Antifreeze Proteins (AFP) and Ice Nucleating Proteins (INP) exemplify nature's adaptation to cold. While AFPs inhibit ice growth, protecting cold-adapted organisms, INPs foster ice formation, counteracting supercooling. Studying these proteins is pivotal for biotechnological advances and understanding nature's thermal strategies.
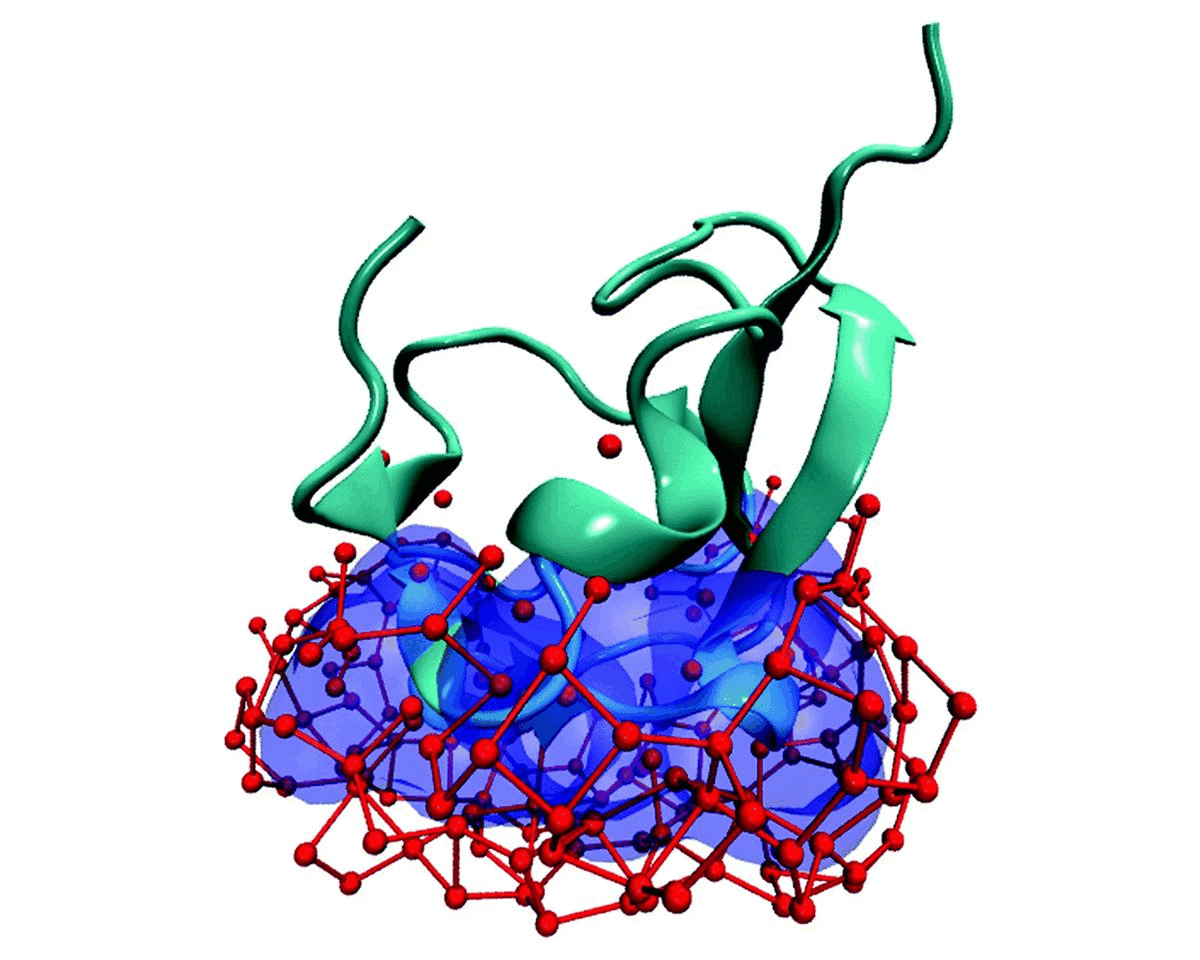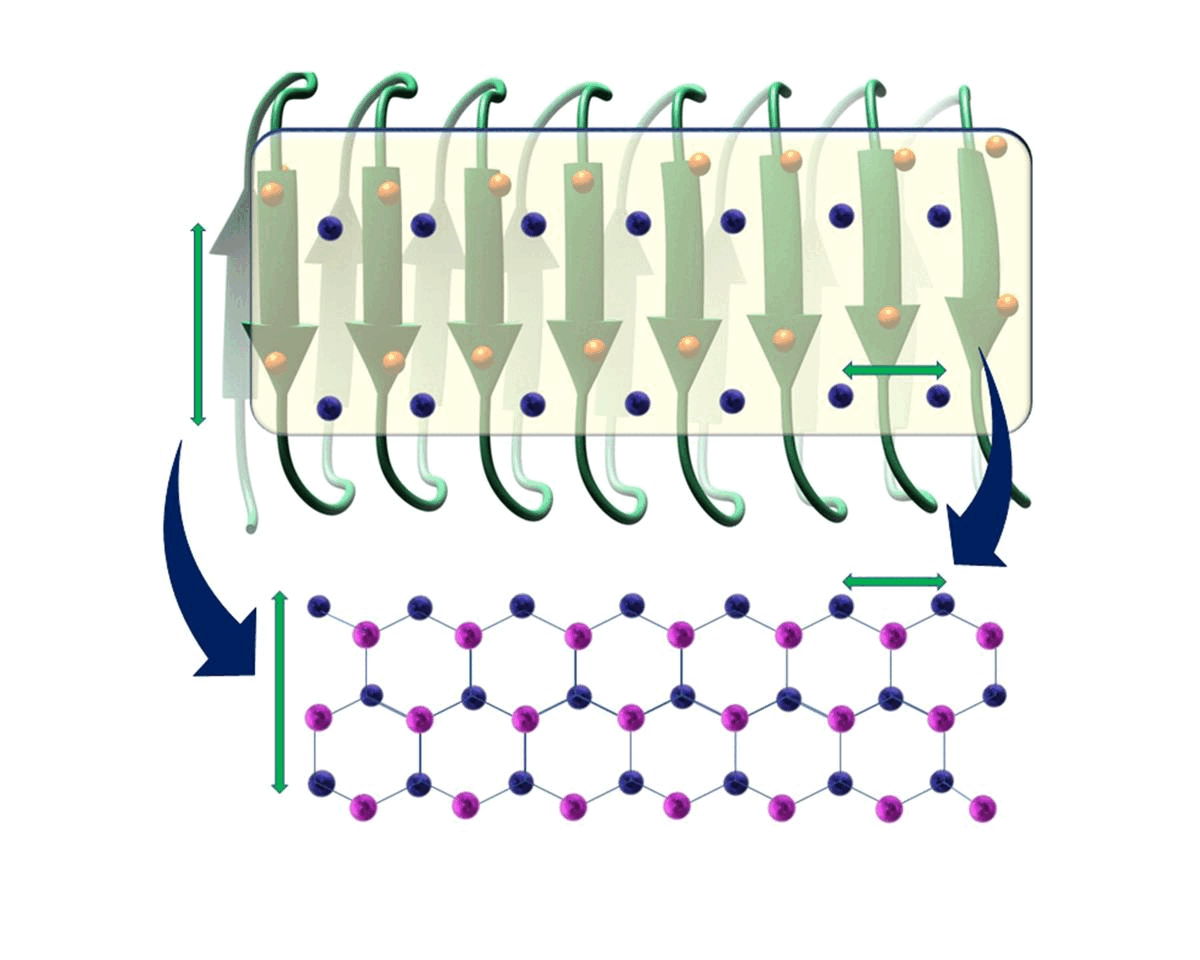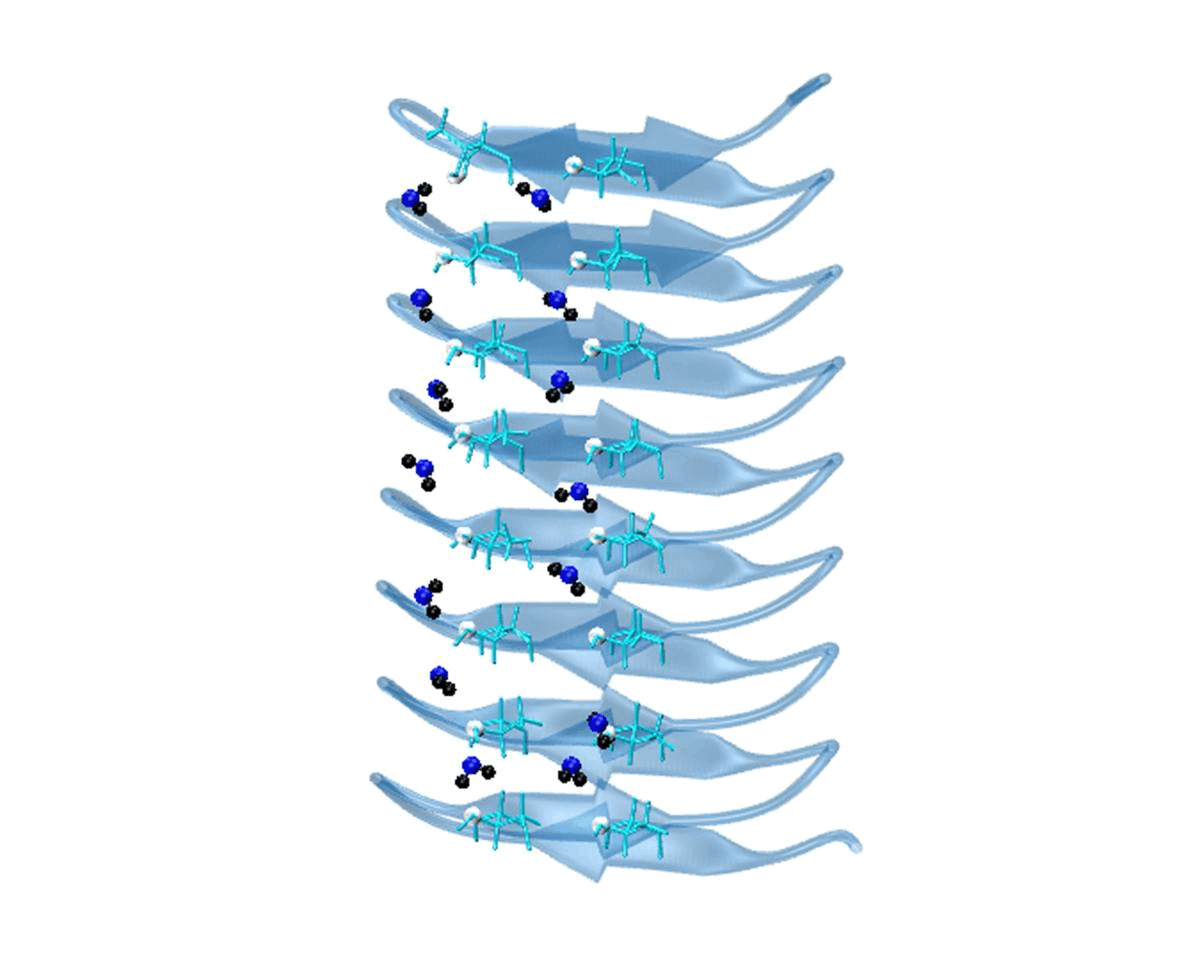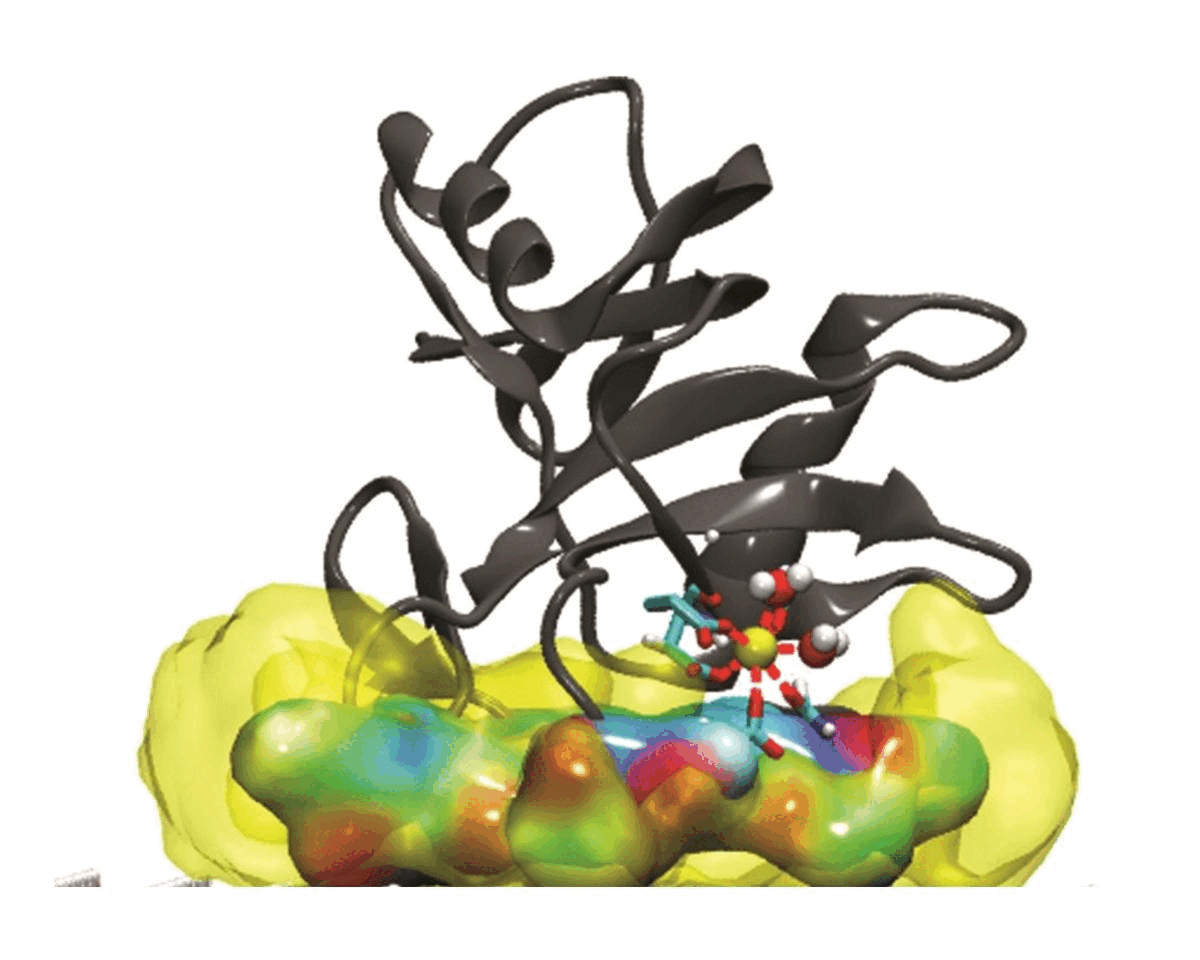
Currently Working: Rahul Aich, Subhabrata Hazra
The Molecular Ballet: How AFPs Inhibit Ice Growth:
Ongoing Work:
Understanding the mechanism of ice growth and growth inhibition by biological and non-biological
substances.
Ice grows uniquely on different planes leading to diverse natural ice formations. Key planes include the basal, prismatic, and secondary prismatic. This growth understanding impacts atmospheric science and cryobiology. Antifreeze Proteins (AFP) in cold-adapted organisms counter ice growth, binding to ice crystals and protecting life in freezing conditions. Deciphering AFP functions offers insights into biophysics and paves the way for biomimetic applications in science.
Currently Working: Rahul Aich, Sourav Satapathi, Sayan Mondal
Constructing free energy surfaces of biological processes using deep learning methods
Ongoing Work:
Currently we are investigating the role of solvent in coil to globule like transition of linear polymer chain and folding kinetics of globular proteins and protein ligand conformational landscape exploration
Flexible polymer chains and proteins can adopt diverse conformations, which exist in different metastable basins in the free energy landscape. Choosing appropriate collective variable for these bio-molecular processes is challenging. Our objective is to obtain the optimized collective variable for protein folding, polymer folding through different linear and non-linear dimensionality reduction techniques. Moreover we are employing markov state model to construct a kinetic network connecting different metastable states.
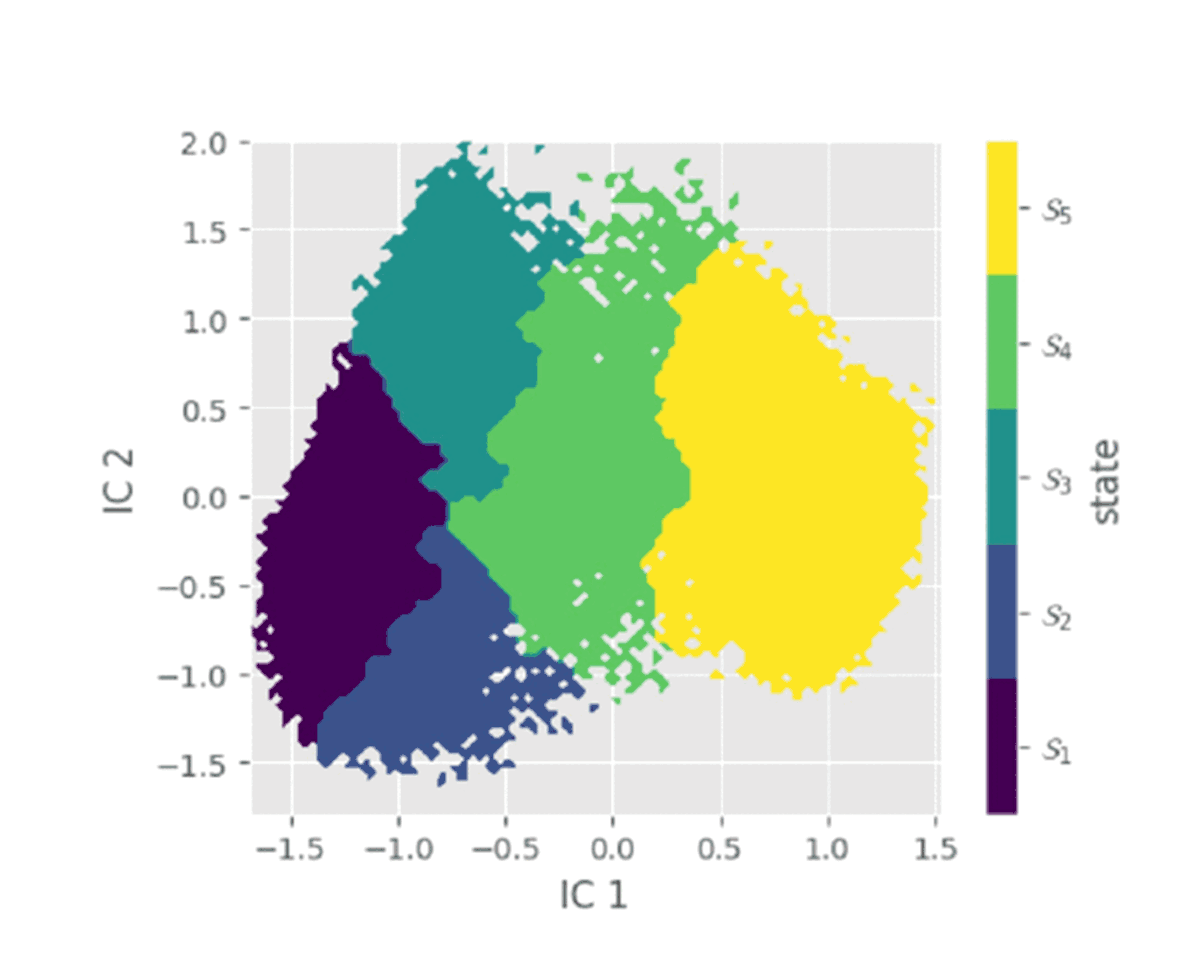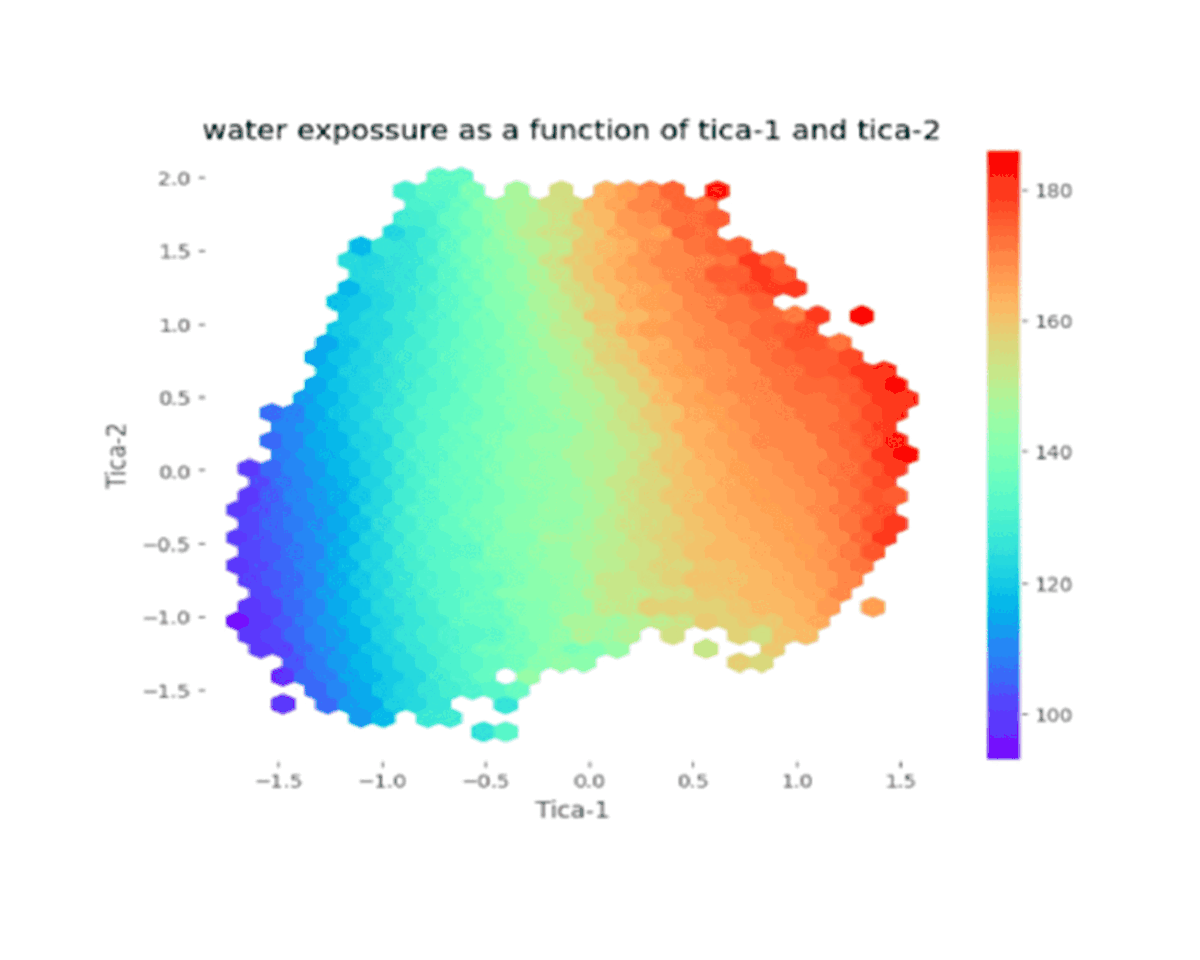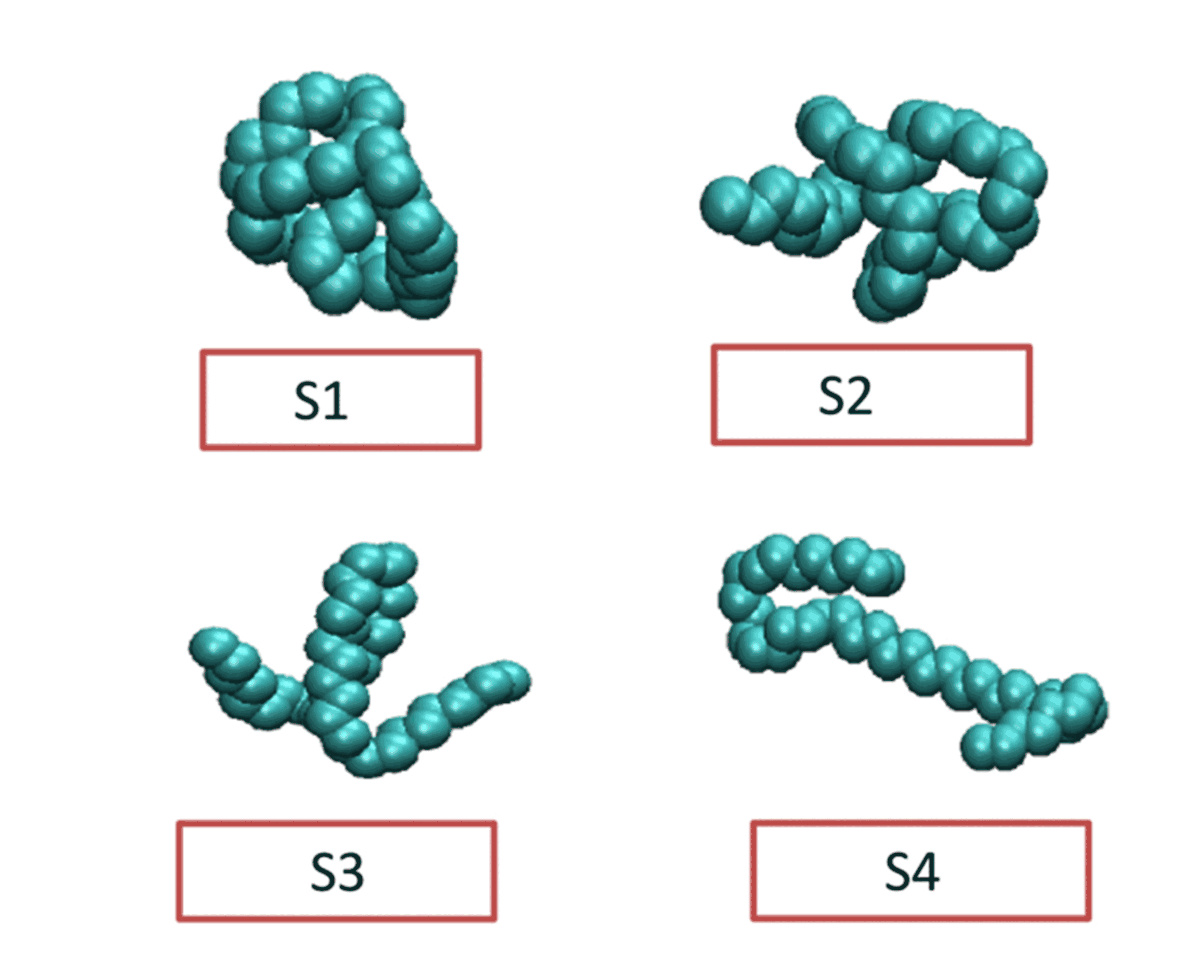
Currently Working: Saikat Dhibar
Theoretical studies of protein folding
Ongoing work
Right now, I'm using math-based models to understand the way proteins fold, focusing on the ones
that don't have a fixed shape (intrinsically disordered proteins). The goal is to see how their unique shape-changing ability
helps them perform various tasks inside cells.
I am focused on applying robust mathematical models to unravel the complexities of biological
processes. My research spans from investigating the fundamental dynamics of protein folding—seeking to understand how
proteins transition from unstructured chains into precise, three-dimensional structures—to exploring the intricate
biological rhythms that govern the internal clocks of various organisms. In both areas, I aim to harness mathematical
theory to quantify these phenomena and shed light on the underlying mechanisms.

Currently Working: Rohon Mitra
Mechanochemical cycle of various Motor Proteins
Ongoing Work:
Currently we are working on the mechano-chemical cycle of dynein motorproteins and their mutational effects
Motor proteins are a class of proteins that use the energy from ATP hydrolysis to move along cytoskeletal filaments within the cell. The process of this mechano-chemical cycle is still not fully understood. However, coarse-grained molecular dynamics studies are being used to uncover the stepping mechanism.

Currently Working: Pradipta Kumar Das
Study of intrinsically disordered proteins
Ongoing Work:
Currently we are working on the folding landscape and coupled folding-binding mechanisms of p53 C-terminal domain
IDPs are proteins that lack a fixed or ordered three-dimensional structure. IDPs are involved in a wide range of cellular processes, including transcription, translation, splicing, signal transduction, and protein folding. We are looking for a generalized description of their folding mechanism and free energy landscape
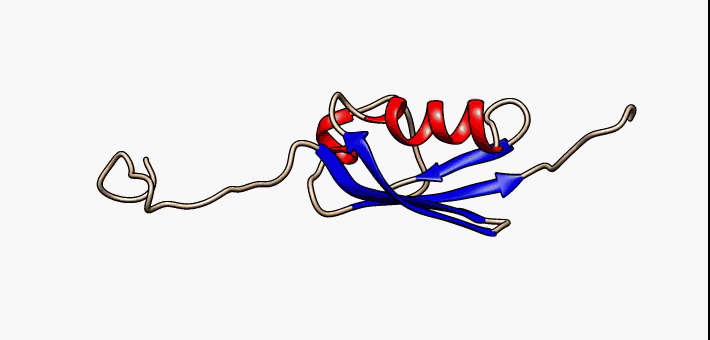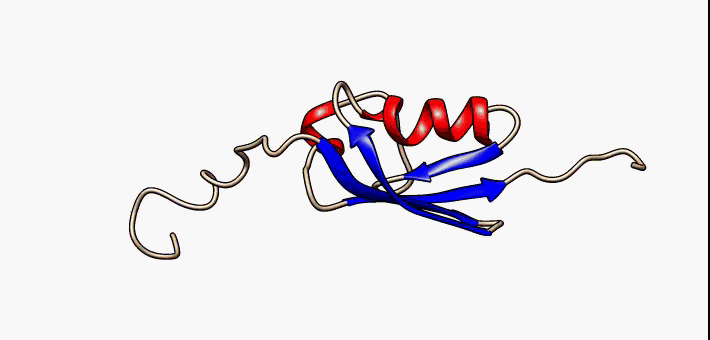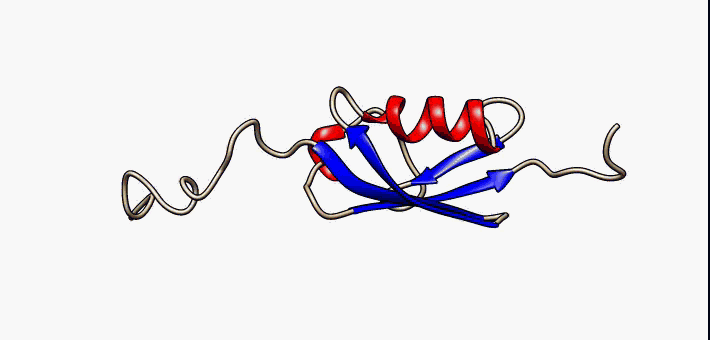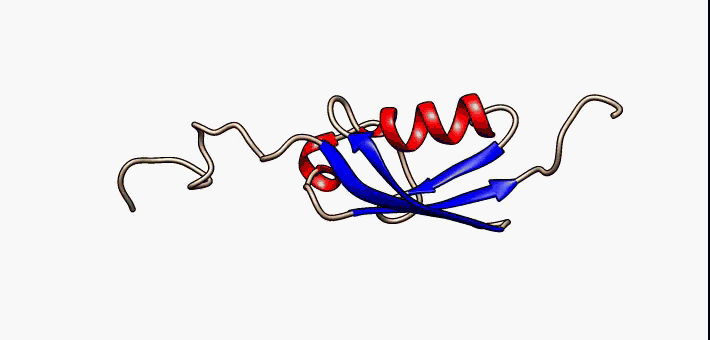
Currently Working: Kingshuk Das
Allosteric Signal Transduction
Ongoing Work: Modern computational tools have revolutionized our understanding of allostery, allowing us to investigate and visualize these intricate interactions. We use Molecular dynamics simulations, in conjunction with network and graph theory approaches which are instrumental in mapping the allosteric pathways, revealing the cryptic corridors of these molecular maestros
Allosteric signal transduction is a delicate dance of molecules, a secret whisper passed from one site to another. It's the tender touch that shifts a protein's shape, setting off a cascade of biological events. This silent symphony of change orchestrates life's functions, turning the key to vibrant vitality. Allosteric signal transduction, with its intricate choreography, forms the very pulse of life's biochemistry. It's nature's master switch, fine-tuning the intricate web of life's processes.
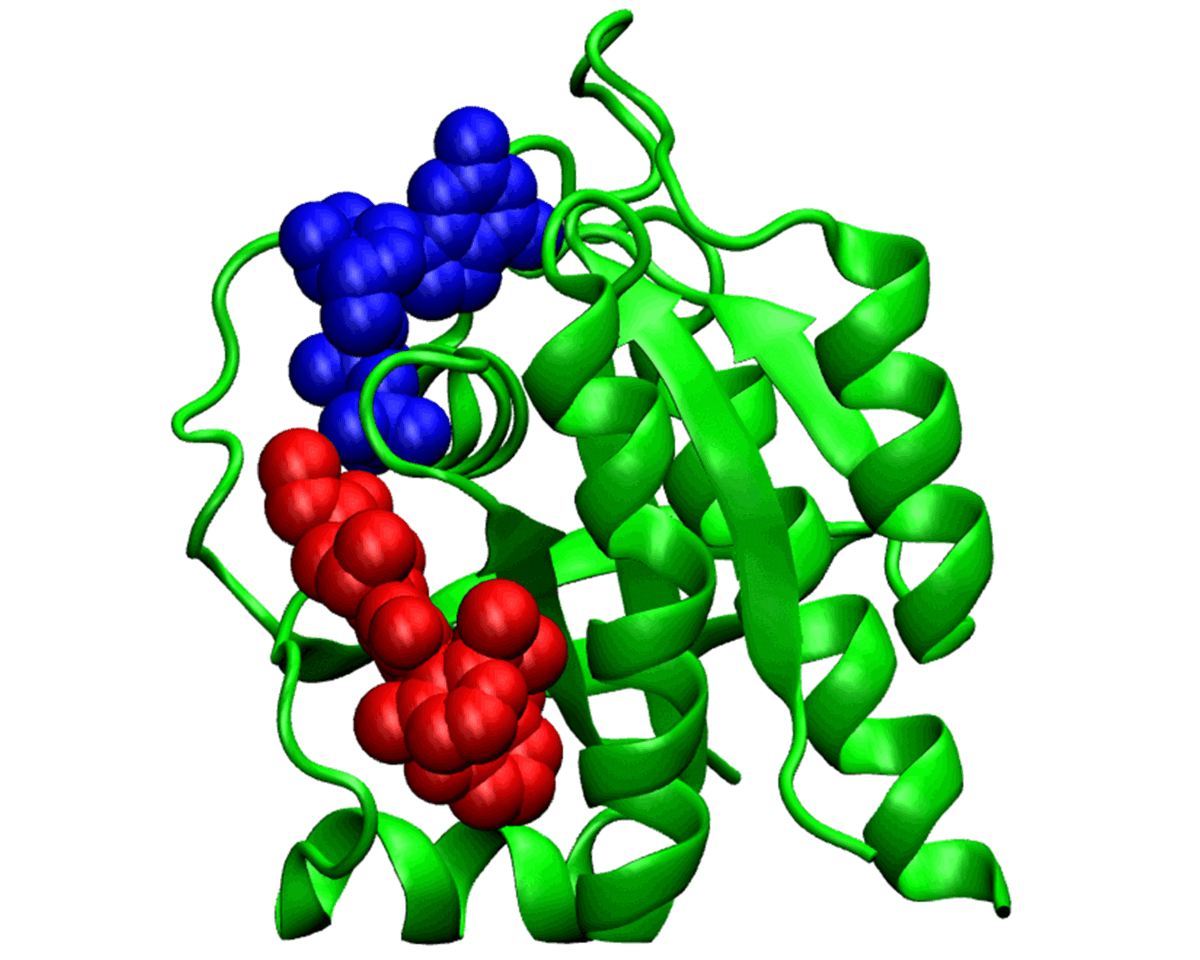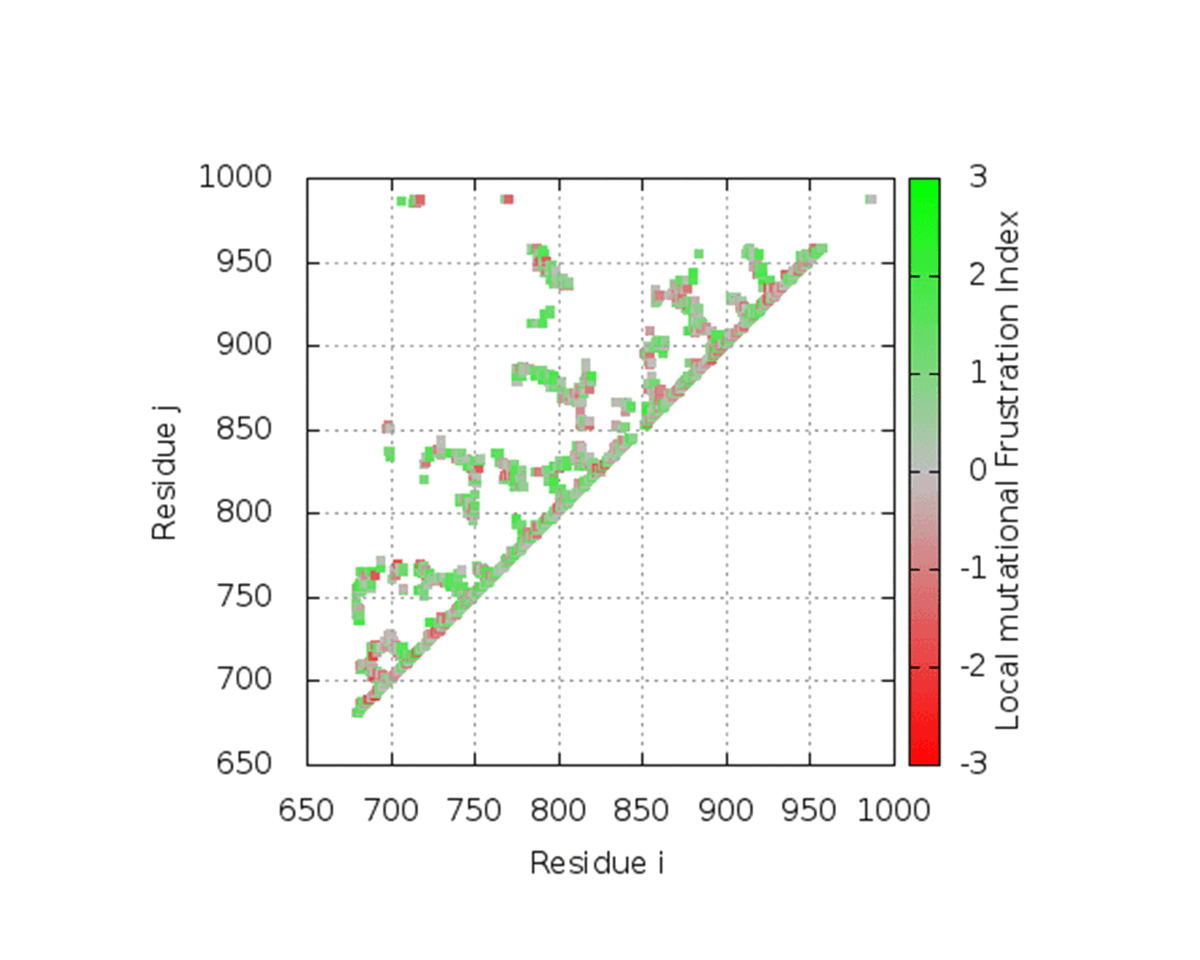
Currently Working: Subhajit Sarkar

